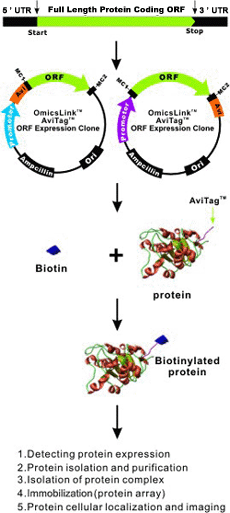
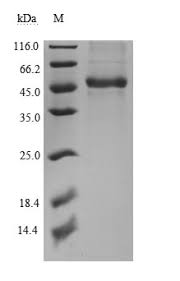
Cusabio Xenopus laevis Recombinant
Abstract
Details of the in vitro synthesis of double-stranded DNA complementary to purified Xenopus globin messenger RNA are presented, using a combination of reverse transcriptase, E. coli DNA polymerase 1 ‘A’ fragment and endonuclease S1. After selection of duplex DNA molecules that approximated the length of Xenopus laevis Recombinant globin messenger RNA by sedimentation of the DNA through neutral sucrose gradients, the 3′-OH ends of the synthetic globin gene sequences were extended. with short stretches of oligo dGMP using terminal transferase.
This material was integrated into linear pCR1 plasmid DNA extended with oligo dCMP and amplified by transfection of E. coli. Plasmids carrying globin sequences were identified by hybridization of 32P-labelled globin mRNA with total cellular DNA in situ, by hybridization of purified plasmids with globin cDNA in solution, by recombinant DNA analysis on polyacrylamide and agarose gels, and by heteroduplex mapping. The results show that extensive DNA copies of Xenopus globin mRNA have been integrated into recombinant plasmids.
Biological source: Xenopus sp.
Quality level: 100
Molecular weight: molecular weight of 17 kDa
Manufacturer/trade name: Upstate®
Technique(s)
activity test: adequate
NCBI Accession Number: NM_002107.3
UniProt access no.: Q16695
Sent in: dry ice
Genetic information: Xenopus laevis…H3F3B(379757)
General description
Product Source: Produced in E. coli.
Quality
Routinely evaluated as a substrate for enzymatic reactions in vitro
Physical form
- HPLC
- Format: Purified
Storage and Stability: 2 years at -20°C
Other notes
For specific activity data, refer to the individual lot Certificate of Analysis for this enzyme.
Legal information
UPSTATE is a registered trademark of Merck KGaA, Darmstadt, Germany
Disclaimer
Unless otherwise stated in our catalogue or other company literature accompanying products, our products are intended for research purposes only and must not be used for any other purpose, including but not limited to unauthorized commercial uses. , in vitro diagnostic uses, ex vivo or in vivo therapeutic uses or any type of consumption or application to humans or animals.
Materials And Methods
- Construction of the expression vector pUC18-PfHly III.
The codon-optimized PfHly III gene (GenScript, Piscataway, NJ; see Fig. S1 in Supplementary Material) was cloned into plasmid pET22b (Novagen-Merck Millipore). PfHly III was amplified from pET22b-PfHly III by PCR with a 5′ primer (5′-GGATCCCATCACCACCATCATCATGAATTCATGGAATTTTACAAAAAC-3′) and a 3′ primer (5′-TCTAGATCAGTGGTGGTTGGTGGTGGTG-3′) to generate BamHI and XbaI sites compatible with plasmid pUC18 (Agilent/Stratagene, Santa Clara, CA). The DNA insert was confirmed by sequencing.
- Bacterial expression of recombinant PfHly III.
The ampicillin-resistant pUC18-PfHly III expression vector was transformed into competent Escherichia coli HB101 cells and cultured at an optical density at 600 nm (OD600) of 0.4, followed by protein induction at 37 °C of 16 h with 1 mM isopropyl-β. -d-thiogalactoside. The bacterial pellet was sonicated in 2 ml phosphate-buffered saline (PBS), followed by microcentrifugation at 12,000 × g at 4 °C.
recPfHly III was purified from the natively soluble supernatant by addition of preloaded Ni2+ nitrilotriacetic acid (NTA) resin and incubated overnight at 4 °C with gentle mixing by end-to-end rotation, followed by centrifugal washes with PBS containing 5 mM imidazole and finally elution with 100 mM EDTA. Elutions were dialyzed overnight at 4°C against PBS (pH 7.5) using Slide-A-Lyzer (Thermo Scientific) to remove EDTA and imidazole. Plasmid pUC18 alone was transformed, induced and purified under the same conditions and used as a negative control.
- Western blot analysis of recPfHly III.
Soluble recPfHly III was separated by SDS-PAGE and transferred to nitrocellulose membrane (Bio-Rad). The membrane was blocked with Qiagen’s blocking buffer at 37 °C for 1 h, washed 3 times with PBS (0.05% Tween 20), and then incubated with a 1:1000 dilution of the anti-His-peroxidase conjugate. horseradish (HRP) on lock. buffer at 4°C overnight. The protein was visualized with enhanced chemiluminescence.
- Hemolytic activity assay.
Human erythrocytes were washed with PBS (pH 7.5) three times and adjusted to a final concentration of 1% (vol/vol). The erythrocyte suspension (0.1 ml) was incubated with 0.1 ml recPfHly III diluted in PBS. The mixture was incubated for a total of 60 min at 37°C. The reaction mixtures were then centrifuged at 1,000 × g for 5 min.
Haemoglobin released from recPfHly III-induced hemolysis was monitored by the absorbance of the supernatant at 550 nm. One hemolytic unit (HU) was defined as the dose of recPfHly III that caused 50% hemolysis. One hundred per cent hemolysis was the amount of haemoglobin released after treating human erythrocytes (1%) with 0.1% Triton X-100. The recPfHly III hemolytic activity assay was studied at the indicated temperatures.
- Osmotic protection experiments.
For osmotic protection experiments, 0.1 ml of recPfHly III (2 HU) was incubated with 0.1 ml of human erythrocytes (1% final concentration) suspended in PBS (pH 7.5) containing an osmotic protector at a final concentration of 30 mM (17). Incubation was at 37°C for 60 min and the mixture was immediately subjected to hemolytic activity assay. Osmotic protectants included glucose, polyethene glycol 600 (PEG 600), PEG 1500, PEG 2000, PEG 3350, PEG 4600, PEG 6000, and PEG 8000 (Sigma). The control used recPfHly III without any osmotic protector.